
In this section, there are a number of examples. The examples go from easy to more sophisticated. Here is a list of the examples and a short description of what you can expect to learn from each of the examples. You can find these examples (and more) in the examples, which is part of the PsyToolkit downloadable software package (see download section).
TIP:
All examples come with the PsyToolkit package. They are in /usr/share/doc/psytoolkit/1.9.4/examples. More examples can be found in the lessons section.
All examples come with the PsyToolkit package. They are in /usr/share/doc/psytoolkit/1.9.4/examples. More examples can be found in the lessons section.
- A simple example. Teaches you:
- How to write a script using an editor
- How to draw a stimulus using Inkscape
- How to compile a script
- How to run an experiment
- How to do some really simple statistics using R
- A Simon task. Teaches you:
- How to set up a simple choice reaction time task
- How to use rectangles as stimuli
- How to save data into a data file
- How to analyze your data with R
- An example of Inhibition of Return. Teaches you:
- How to set up a choice reaction time experiment with multiple stimuli in one trial
- A more advanced example of Inhibition of Return. Teaches you:
- How to use R for creating feedback for participants
- A example of task switching. Teaches you:
- How to use multiple tasks
- An example of using the mouse as a response devices. Teaches you:
- How to use the mouse in scripts
- How to ask the participant to click on certain areas (stimuli) of the screen
- How to use fonts
- An implementation example of the Wisconsin Card Sorting Test. Teaches you:
- How to identify which bitmaps a participant clicked
- Set and use a custom variable
- Run a fixed sequence of trials
- How to use the pager function to show multiple instruction screens
- Play sounds
- An implementation example of an attentional blink task. Teaches you:
- How to run experiments with or without a table
- How to clear multiple stimuli in one line
- An implementation example of a multiple object tracking task task. Teaches you:
- How to use moving stimuli sprites
- How to click moving stimuli with the mouse
- How to show a background image
TIP:
These examples can all be found in the downloadable PsyToolkit package!
These examples can all be found in the downloadable PsyToolkit package!
A complete guide for a simple experiment
Before we start with anything advanced, I would like to take you through a complete example of a simple experiment. It may help to get used to the idea of PsyToolkit, whilst leaving out more advanced features. I will also show how to analyze the data with the free statistical software R. This is just to give you a start and a feel for the whole approach of PsyToolkit. If you got that, you can move to the other examples further down on this page.Step 1: Design
The experiment will have 10 trials of a simple task. The task is: As soon as a red circle appears, the participant must press the space bar. After this button press, the circle appears, and will appear soon after for a next trial. The maximum allowed response time is 1 second.Step 2: Create a new directory
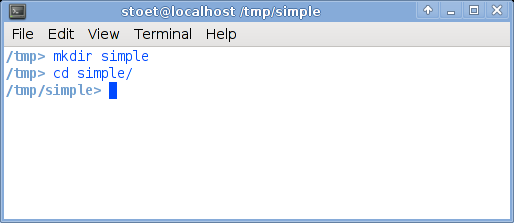
It is a good idea to create a separate directory (or "folder") for each experiment. In Linux, you can create directories either on the Command Linux Interface, or using the graphical file manager. Here, I will explain the command-line way (because you can figure the point-and-click way yourself).- Open a 'terminal window'. You can do this, on many Linux systems as follows: Go to "applications", "accessories", "terminal".
- Create a directory called simple, type mkdir simple.
- Go to this directory typing cd simple.
TIP:
Typing things into the terminal is known as a command line interface. Read more about this for Linux in this guide.
Typing things into the terminal is known as a command line interface. Read more about this for Linux in this guide.
Step 3: Create a stimulus
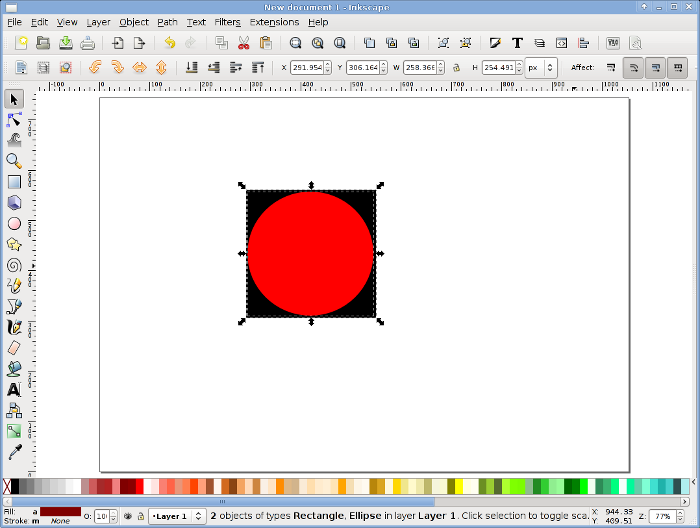
For this experiment, we need only one stimulus, a red circle. Create a red circle on a black rectangle. This way, on the black screen (the default color in PsyToolkit), you will just see a red circle.- Open Inkscape, either from the menu or by typing Inkscape on the command line.
- Draw a stimulus, for this experiment is does not really matter what it looks like. For example, click on the circle box in the left toolbox column and then draw a circle. You can fill it in a different way by clicking the right mouse button on the drawn circle and select fill and stroke. The Inkscape user interface is intuitive. When you are done with drawing the stimulus, select it by clicking on it or drawing a selection box around it.
- In Inkscape, select from the File menu the export bitmap option. You will see a dialog window. Name the bitmap mystimulus.png and click the export button (make sure you choose the right directory as well, and set the width not too big, for example 150 by 150 pixels). Now there should be a bitmap file in png format in your directory!
- Exit the Inkscape program now (but you might save the SVG file as well, the graphics file format of Inkscape, even though PsyToolkit does not use SVG files itself).
- Check whether your new file mystimulus.png exists in the correct directory. Just type ls in the terminal window to get a list of all your files in the current directory. If it does not exist, you did something wrong, and need to try Inkscape again.
You are done with the stimulus creation. If you want to learn more of how to use Inkscape, there is a good online book on how to use it: tutorial.
TIP:
Stimuli can be transparent (but that is not the default option). With transparency, you do not need to put stimuli on a black background (as in the above example). All you need to do is choose the transparency option: transparency on. A good example is the first image in the multiple object tracking task, where the faces are semi transparent (you can see the clouds through them).
Stimuli can be transparent (but that is not the default option). With transparency, you do not need to put stimuli on a black background (as in the above example). All you need to do is choose the transparency option: transparency on. A good example is the first image in the multiple object tracking task, where the faces are semi transparent (you can see the clouds through them).
Step 4: Open up an editor and write a simple script
Now you need to write your PsyToolkit script file. All this file is, is some text. You can create such a file with a text editor. Maybe you already have a favorite text editor. If not, and if you are not very familiar with text editors, you can use gedit, which is a sophisticated text editor installed on many Linux systems. Often you can select this from your desktop menu: Applications->Accessories->Text Editor. Or simply type gedit on the command line.Open editor and copy and paste this code
bitmaps mystimulus task simpletask keys space delay 100 show bitmap mystimulus readkey 1 1000 clear 1 save STATUS RT block firstblock tasklist simpletask 10 end
Now the same, but with full explanation of each line
# everything following "#" is comment
bitmaps # indicates definition of bitmaps
mystimulus # refers to the file mystimulus.png
# empty line means end of (bitmaps) section
task simpletask # task indicates definition of the "simpletask"
keys space # there is only one key used, the space bar
delay 100 # wait for 100 milliseconds
show bitmap mystimulus # show the bitmap. Without XY coordinates show at center
readkey 1 1000 # wait up to 1000 milliseconds for key number 1
clear 1 # once key pressed (or 1000 ms passed), clear first presented stimulus
save STATUS RT # save the status (correct/timeout) and reaction time to file
block firstblock # blocks tell how to run trials
tasklist # lists which tasks to choose from
simpletask 10 # run simpletask 10 times
end # end of task list
In gedit, it will look like this:
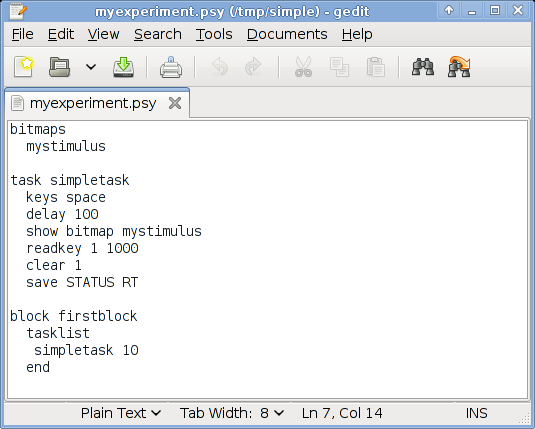
Copy and paste this in your editor. Save the file as myexperiment.psy. Make sure you save it in the right directory (that is the one you created before)! You can close the text editor, or leave it open, as you like.
As you can conclude from the example, the code consists of three parts, one describes the stimuli, one describes the task, and one describes the experimental block. Of course, you can have multiple tasks and blocks, etc, but here I just want to give a possibly simple example.
Step 5: Compile the script
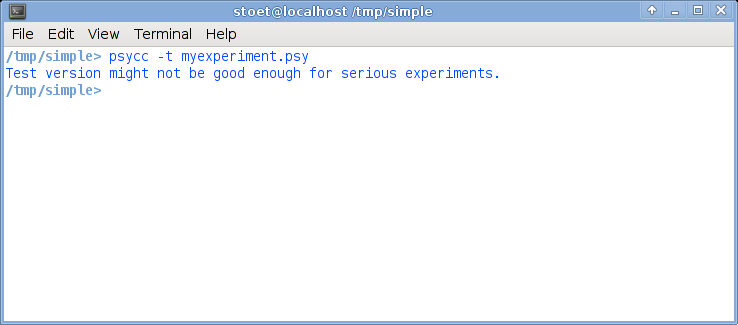
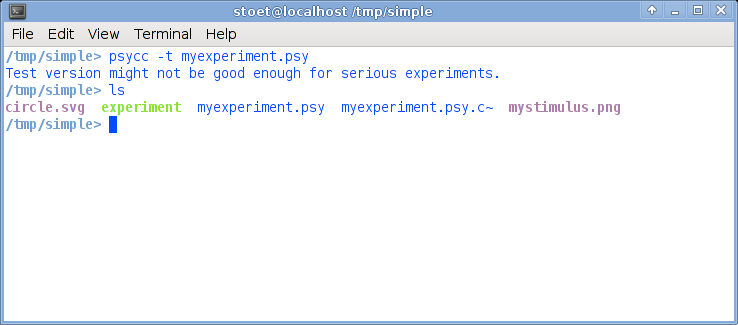
Now select your terminal window again. Now type psycc -t myexperiment.psy, obviously followed by return. The -t option tells the compiler that you just want to test, and it will run in a window, not in full screen mode.This is what it could look like:
Just for testing: List the contents of the directory (type ls to get a listing of all files in the directory). There should now be a file called experiment.
This is what it could look like:
Step 6: Test the experiment
On the command line type experiment. Nothing happens? Than the current path is probably not in your path variable. Try ./experiment, that should work for sure. You should now get a full screen experiment with 10 trials. If you see the dot, just press the space bar.This is what it could look like:
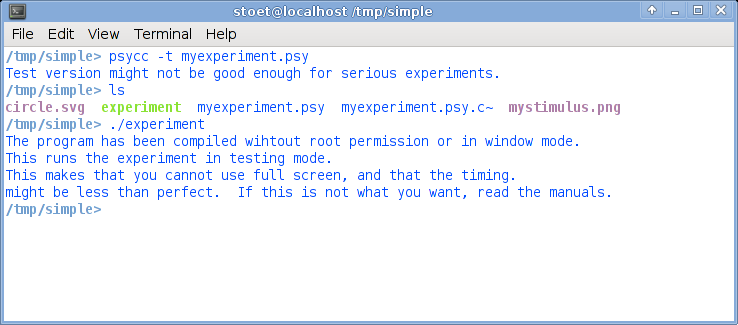
With the option -t, psycc runs in a testmode. If you want to run in regular mode, you will not get all of the messages you see now. But, because we are testing here, it is good to work in test mode. Also, when you run in testmode, you will not run in full screen mode.
TIP:
Compile with the -t option to test scripts. Once it works, you compile without -t to run in full screen mode.
Compile with the -t option to test scripts. Once it works, you compile without -t to run in full screen mode.
Step 7: Analyze the data
Now you are done and a new file should have been created, something starting with myexperiment and ending with data. Just type ls to find the new file, and type cat plus data filename to see the content (you can also see and edit the content with gedit, of course).This is what it could look like:
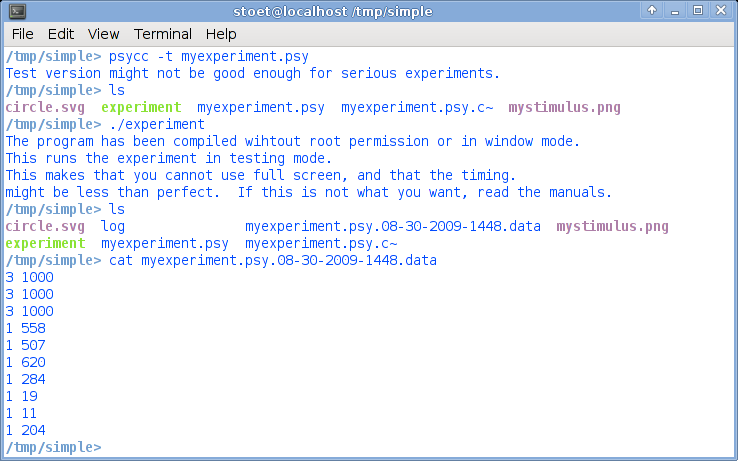
As you can see, the data file contains the date (you can open it with the text editor, it is a simple text file, with one line for each trial). Write down the file name on a piece of paper.
The first column is the status (1=correct, 3=timeout), and the second column is the time in ms. If you would never have pressed the button, all lines would have been "3 1000". For this example, I did not press the first 3 trials. Of course, you can set the maximum RT from 1000 ms to something longer.
Now type R (assuming you have installed the package R). You will get a prompt. Type the following, but replace the "DATAFILE" with the filename you just wrote down. You will get the mean response time!
data = read.table("DATAFILE")
meanrt = mean( data[ data[,1]==1 ,2] )
cat("Mean response time in correct trials is:", meanrt,"\n")
q()
That was the simple example!
More advanced examples
Before you move on, please note that the example files are located in /usr/share/psytoolkit/examples/You can only compile an example if you first copy it to a write permissible location. For example, if you want to test the simple experiment yourself, and if you are in your home directory, you can type this:
Compile and run an example experiment.
# Type the following on the command line, assuming the $ sign # is your command line prompt: $ cp -r /usr/share/psytoolkit/exampels/simple ./ $ cd simple $ psycc -t -k simple.psy $ ./experiment
Simon task, also known as spatial compatibility task
In short, compatibility tasks study the ease of difficulty of the relationship between stimuli and responses, stimuli and stimuli, or responses and responses. Examples of incompatible relationships are:- Drawing a circle with your left hand whilst drawing a rectangle with your right hand
- Responding with your left hand to something happening right of you.
Some of the early work on compatibility was done by J.R. Simon.
In the spatial compatibility task of the example, you must respond to red and green stimuli with the left shift or right shift key, respectively. If the red stimulus appears on the left side of the screen, it is easier to respond to than when it appears on the ride side of the screen.
In this example, no bitmaps are used. The aim of the example is to show how you can create simple stimuli using rectangles.
Another aim is to show positioning of stimuli. Each stimulus has an X and a Y coordinate. If you use the option centerzero, XY coordinates 0,0 refer to the screen center. Thus, if you present a stimulus at XY position -200 0 , the stimulus will be horizontally left of the screen center, and vertically in the middle. In the simon task, we present the fixation point at screen center (0,0), and the target stimuli left or right of the center.
Compile this experiment just with psycc -t simon.psy. Run the experiment by typing ./experiment. After the experiment, type ls -rtl in your terminal window, and see that some new files have been created. One of these files ends with the extension data. Open this file with a text editor. Each row of this file has the data of one trial. Each row is created by the save line of the script.
# File: examples/simon-spatial-compatibility/simon.psy
# simple Simon stimulus-response compatibility task
# instruction for participant:
# press left shift button in response to red stimulus
# press right shift button in response to green stimulus
options
centerzero # postion 0,0 is center of screen
window # use a window (remove this to have full screen)
escape # experiment can be interupter by holding escape until end of trial
table simontasktable # table with four rows, each trial one row is randomly chosen
#-name of condition--------------position--red?--green?--correct key--------------
"leftresponse compatible" -200 255 0 1
"leftresponse incompatible" 200 255 0 1
"rightresponse compatible" 200 0 255 2
"rightresponse incompatible" -200 0 255 2
task simon
table simontasktable # table to be used
keys lshift rshift # keys to be used
show rectangle 0 0 10 10 255 255 255 # small white rectangle at
delay 200 # center for 200 ms
clear 1 # erase the rectangle
delay 50 # and wait 50 ms
show rectangle @2 0 100 100 @3 @4 0 # target stimulus is presented
readkey @5 5000 # wait for key of column 5 of table
clear 2 # remove target stimulus from screen
if STATUS != CORRECT
show rectangle 0 0 10 10 255 255 255
delay 100
show rectangle 0 0 20 20 255 255 0
delay 100
show rectangle 0 0 30 50 255 255 255
delay 100
show rectangle 0 0 50 50 255 255 0
delay 100
clear 3 4 5 6
fi
delay 100 # wait 100ms (here sort of intertrial time)
save BLOCKNAME @1 TABLEROW KEY STATUS RT # save data to file
block test # this block is called "test"
tasklist
simon 40 # run the simon task 40 trials.
end
Run the experiment and analyze the data with the following R file. Here is how you can do that:
- Run gedit from command line with gedit, and copy and paste the code below.
- Replace the filename on the read.table line with your data filename.
- Save file as analyze.r and exit gedit.
- Run R, just type R on the command line.
- On the R prompt, type source "analyze.r".
- You should now see a box plot.
# File: examples/simon-spatial-compatibility/analyze.r
# read the table. Filename has date of finishing time in it, so
# it will be different on your computer. Look up which file it is using
# ls simon*data
# on the command line.
data = read.table( "example.data" )
# boolean, True if trial was correct
correct = data[,6]==1
# boolean. True if compatible trial
compatible = data[,3]=="compatible"
# boolean. True if response was on the left
left = data[,2]=="leftresponse"
# rt is in column 7, that is data[,7]
# use cbind to combine the data vectors of the four conditions
boxplot( cbind( data[ correct & compatible & left, 7],
data[ correct & compatible & !left, 7],
data[ correct & !compatible & left, 7],
data[ correct & !compatible & !left, 7]),
names=c("left/comp","right/comp","left/incomp","right/incomp"),
ylab="RT (ms)",col=c(3,3,2,2))
title("Box plots of RTs in the four conditions")
It might not be the case that you were slower in the incompatible trials (in red). Stimulus-response time compatibility effects can be small. You could change the number of trials to something like 500, and do the analysis again.
Inhibition Of Return
Inhibition Of Return is the phenomenon that people respond slower to a stimulus which occurs at the same position as a target. The phenomenon was first described by Michael Posner.In the following experiment, we have the following sequence of events:
- Two yellow frames, left and right of a fixation cross are simultaneously presented
- One of the two frames flashes. This should attract the attention to either the left of the right. This is called the IOR Cue.
- About a second later, within one of the two frames, a white dot appears (i.e., the IOR target), which requires an immediate left or right button press (left or right shift button)
Code: Creating a simple IOR experiment
# File: examples/ior/ior.psy # example script doing an inhibition of return experiment # everything following a hash mark is a comment and is ignored # by the script interpreter options window origin topleft escape # you can escape the program render doublebuffer bitmaps # define some bitmap names, different ways to do this. frame bitmaps/frame.png plus bitmaps/plus.png dot bitmaps/dot.png instruction bitmaps/instruction.png table standard # table with 5 columns, which can be address with @-sign #1 #2 #3 #4 #5 #-------------------------------------------- "cueleft targetleft cued" 2 200 200 1 "cueleft targetright uncued" 2 200 600 2 "cueright targetleft uncued" 3 600 200 1 "cueright targetright cued" 3 600 600 2 task ior table standard # use this table for choosing conditions keys lshift rshift # use 2 shift keys in this experiment draw off # hold off drawing show bitmap plus # 1 show bitmap frame 200 300 # 2 show bitmap frame 600 300 # 3 draw on # now draw all three items delay 1500 # wait 1500 ms clear @2 # IOR cue; frame OFF attract attention delay 100 # wait 100 ms show bitmap frame @3 300 # 4 ; frame ON again, still attacts attention delay 860 # wait 860 ms show bitmap dot @4 300 # 5 ; show IOR target readkey @5 1500 # wait max 1500 ms until correct key is pressed clear 5 # remove target from screen save BLOCKNAME @1 STATUS RT # save trial information message instruction # shows instruction and waits for key press block mainblock # there is just one block tasklist ior 10 # choose the ior task 100 times end
Note that I like indentations. They are only used for readability, they have no meaning to the interpreter. On the other hand, EMPTY LINES play a syntactical role. An empty line ends a section, such as the sections 'options', 'bitmaps', 'table', 'task', and 'block'.
Make a directory and put this code in a textfile (or go to the corresponding examples directory in the downloaded package). Get this code running by typing the following on the command line. The "$linux" part is assuming to be the prompt, this will likely be different on each system.
Compile and run
$linux> psycc ior.psy password: your password, make sure you are in the sudo group $linux> ./experiment
The options part has the option window. Take it out to get the default full screen mode!
Compile and run, method 2
$linux> psycc ior.psy $linux> experiment
Now do the experiment. The data will be written to a file starting with the name of the script, and ending in data. Every data file will be unique, since it contains the date and time of the time it gets written (at the end of the data collection).
Of course, you want to analyze the data. You can do it with whatever program you like, the data is save in just plain ASCII. Here are some examples of how you can do it in R. Of course, R is a whole language on itself, and you can find great tutorials on the internet (see the resources section).
R Example: Analyze the data of a single subject
# File: examples/ior/ior-single-subject.r
files = dir(pattern="^ior\.psy.*data$")
if ( length(files) > 1 )
{
cat("there is more than one datafile, I will look at the first one\n")
}
data = read.table( files[1] )
colnames(data)=c("block","cuepos","targetpos","condition","status","rt")
rt = data[,"rt"]
correct = data[,"status"]==1
cued = data[,"condition"]=="cued"
cat("Cued mean: ",
mean(rt[correct & cued]),"\n",
"Uncued mean: ",
mean(rt[correct & !cued]),"\n",sep="")
R Example: Analyze the data of a group of subjects with paired Student's t tests
# File: examples/ior/ior-group.r
# assume all files ending in .data are from this experiment!
cued.all=numeric(0)
uncued.all=numeric(0)
datafiles=dir(pattern="data$")
cat("Number of subjects: ",length(datafiles),"\n")
if ( length(datafiles) < 2 ){cat("not enough data!\n")}else{
for ( i in datafiles )
{
x=read.table(i)
cued = mean(x[x[,4]=="cued" & x[,5]==1,6])
uncued = mean(x[x[,4]=="uncued" & x[,5]==1,6])
cued.all = append( cued.all, cued )
uncued.all = append( uncued.all, uncued )
}
cat("Cued mean: ",mean(cued.all),"\n","Uncued mean: ",
mean(uncued.all),"\n",sep="")
print(t.test(cued.all,uncued.all,paired=T))
}
Inhibition Of Return, but fancier.
Probably you want some things look a little nicer. For example, you might want one training block, and you want blocks with pauses between them, and you would like your subjects to be motivated, so you might want to give them some feedback about their performance. The following example has all those features. Note that you can ask a block to repeat itself by adding a number in the block line. Also not that you can make a system call to access the data. In this case, following every block, we call R to analyze the data of the block and then display the results.The feedback screen looks like this:
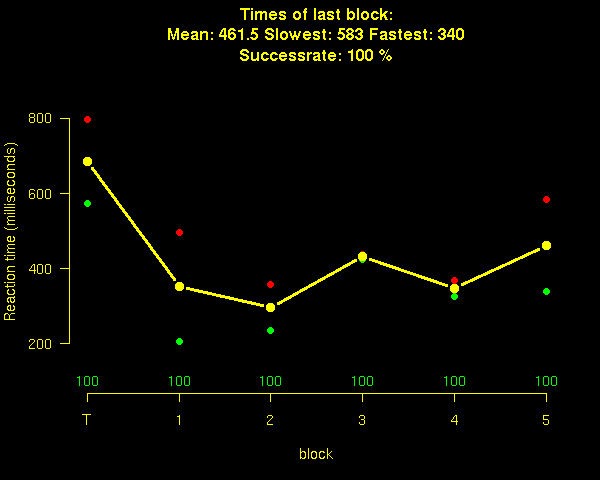
code: ior fancy version
# File: examples/ior/ior2.psy # example script doing an inhibition of return experiment # everything following a hash mark is a comment and is ignored # by the script interpreter options window # remove this if you want to run full screen escape # you can escape the program bitmapdir bitmaps # directory where to find bitmaps origin topleft bitmaps # define some bitmap names frame # if bitmaps have png extension, you do not need to specify plus # the whole file name. dot instruction table standard # table with 5 columns, which can be address with @-sign #1 #2 #3 #4 #5 #-------------------------------------------- "cueleft targetleft cued" 2 200 200 1 "cueleft targetright uncued" 2 200 600 2 "cueright targetleft uncued" 3 600 200 1 "cueright targetright cued" 3 600 600 2 task ior table standard # use this table for choosing conditions keys lshift rshift # use 2 shift keys in this experiment draw off # hold off drawing show bitmap plus # 1 show bitmap frame 200 300 # 2 show bitmap frame 600 300 # 3 draw on # now draw all three items delay 1500 # wait 1500 ms clear @2 # IOR cue; frame OFF attract attention delay 100 # wait 100 ms show bitmap frame @3 300 # 4 ; frame ON again, still attacts attention delay 860 # wait 860 ms show bitmap dot @4 300 # 5 ; show IOR target readkey @5 1500 # wait max 1500 ms until correct key is pressed clear 5 # remove target from screen save BLOCKNAME @1 STATUS RT # save trial information message instruction # show instruction, and wait for space key press block training # training block tasklist ior 2 # choose the ior task 5 times end system R CMD BATCH create-feedback.r bitmap_from_file feedback.png wait_for_key block main 5 # 5 blocks of 10 trials tasklist ior 2 # choose the ior task 5 times end system R CMD BATCH create-feedback.r bitmap_from_file feedback.png wait_for_key
R script to have in the same directory.
# File: examples/ior/create-feedback.r
x = read.table("/dev/shm/data")
postscript(file="feedback.ps",width=600,height=480)
mins = rep(0,6)
maxs = rep(0,6)
mns = rep(0,6)
error= rep(0,6)
BLOCK = as.character(x[,1])
RT1 = x[,6]
blocks = unique(BLOCK)
CORRECT = x[,5] == 1
data = numeric(0)
for ( i in 1:length(blocks) )
{
mns[i] = round(mean(RT1[ CORRECT & BLOCK==blocks[i] ]),digits=1)
mins[i] = round(min(RT1[ CORRECT & BLOCK==blocks[i] ]),digits=1)
maxs[i] = round(max(RT1[ CORRECT & BLOCK==blocks[i] ]),digits=1)
error[i]= round(
sum(CORRECT & BLOCK==blocks[i])/sum(BLOCK==blocks[i])*100,digits=1)
}
par(bg="black",fg="yellow",col.main="yellow",col.axis="yellow",col.lab="yellow")
plot(0,0,xlim=c(1,6),ylim=c(100,max(maxs)+100),axes=F,
ylab="Reaction time (milliseconds)",xlab="block")
title(paste("Times of last block:\n Mean:",mns[length(blocks)],"Slowest:",
maxs[length(blocks)],"Fastest:",mins[length(blocks)],
"\nSuccessrate:",error[length(blocks)],"%"),col="yellow")
points(mins[1:length(blocks)],pch=19,col="green")
points(maxs[1:length(blocks)],pch=19,col="red")
points(mns[1:length(blocks)],pch=19,col="yellow",type="b",lwd=3)
for ( i in 1:length(blocks))
{
color = "green"
if ( error[i] < 90 ){color="orange"}
if ( error[i] < 80 ){color="red"}
text( i , 100 , error[i],col=color)
}
labs=c("T",paste(1:5,sep=""))
axis(1,at=1:6,labels=labs)
axis(2,las=1)
dev.off()
system("convert -rotate 90 feedback.ps feedback.png")
Task switching
Task switching paradigms are inspired by the work of Jersild (1927), much later refined and implemented as computerized tasks. There are hundreds of studies using task switching paradigms.In task-switching paradigms, subjects usually perform two tasks. Each trial starts with a task cue, telling which of the two tasks to do. Then they apply the task rule to the upcoming stimulus. Switch costs occur when subjects switch between tasks.
In the example script, a color and a shape task are randomly interleaved. Every trial starts with a task cue, followed by a stimulus. In the color task, red requires a left and green requires a right button press. In the shape task, a circle requires a left and a square a right button press.
In this script, I have two "tasks", one for each of the tasks of the paradigm, and I have two "tables".
Script: Creating a simple taskswitching experiment
# File: examples/taskswitching/simple/taskswitching.psy
options
window
bitmaps
colorcue bitmaps/colorcue.png
shapecue bitmaps/shapecue.png
red_circle bitmaps/redcircle.png
green_circle bitmaps/greencircle.png
red_square bitmaps/redsquare.png
green_square bitmaps/greensquare.png
ready bitmaps/ready.png
thanks bitmaps/thanks.png
sounds
error sounds/wrong.wav
table colortask
#1 2 3
#-----------------------------------------
"color congruent left " red_circle 1
"color incongruent right" green_circle 2
"color incongruent left " red_square 1
"color congruent right " green_square 2
table shapetask
#1 2 3
#-----------------------------------------
"shape congruent left " red_circle 1
"shape incongruent left " green_circle 1
"shape incongruent right" red_square 2
"shape congruent right " green_square 2
task color
table colortask
keys lshift rshift
show bitmap colorcue # stimulus 1
delay 1500
clear 1 # clear stimulus 1
delay 400
show bitmap @2 # stimulus 2
readkey @3 2000
clear 2 # clear stimulus 2
if STATUS == WRONG
sound error
delay 2000
fi
delay 1000
save @1 STATUS RT
task shape
table shapetask
keys lshift rshift
show bitmap shapecue # stimulus 1
delay 1500
clear 1 # clear stimulus 1
delay 400
show bitmap @2 # stimulus 2
readkey @3 2000
clear 2 # clear stimulus 2
if STATUS == WRONG
sound error
delay 2000
fi
delay 1000
save @1 STATUS RT
block mainblock
bitmap ready # shows "ready?"
wait_for_key # wait for pressing the space bar
tasklist # random interleave
color 10 # 10x color task
shape 10 # 10x shape task
end # total 20 trials
bitmap thanks # shows "thanks!"
wait_for_key # wait for pressing the space bar
R Example: Analyze single subject
# File: examples/taskswitching/simple/taskswitching-single-subject.r
files = dir(pattern="^taskswitching\.psy.*data$")
if ( length(files) > 1 )
{
cat("there is more than one datafile, I will look at the first one\n")
}
data = read.table( files[1] )
colnames(data)=c("task","congruency","response","status","rt")
rt = data[,"rt"]
correct = data[,"status"]==1
repetition=c(F,data[1:length(data[,1])-1,"task"]==data[2:length(data[,1]),"task"])
congruent=data[,"congruency"]=="congruent"
options(digits=2)
cat("switch cost (switch - repeat):",mean(data[correct & !repetition,"rt"])-
mean(data[correct & repetition,"rt"]) , "ms\n")
cat("task interference (incongruent - congruent):",mean(data[correct & !congruent,"rt"])-
mean(data[correct & congruent,"rt"]) ,"ms\n")
R Example: Analyze group subject with repeated measures ANOVA
# File: examples/taskswitching/simple/taskswitching-anova.r
files = dir(pattern="^taskswitching\.psy.*data$")
if ( length(files) < 2 ){cat("not enough data\n")}else{
alldata = numeric(0)
subject = character(0)
switchcon = character(0)
congruency= character(0)
subjectnumber=1
for ( i in files )
{
# analyze data of one subject
data = read.table( i )
colnames(data)=c("task","congruency","response","status","rt")
rt = data[,"rt"]
correct = data[,"status"]==1
repetition=c(F,data[1:length(data[,1])-1,"task"]==data[2:length(data[,1]),"task"])
congruent=data[,"congruency"]=="congruent"
options(digits=2)
switchconmean = mean( rt[correct & !repetition & congruent ])
repeatconmean = mean( rt[correct & repetition & congruent ])
switchincmean = mean( rt[correct & !repetition & !congruent])
repeatincmean = mean( rt[correct & repetition & !congruent])
alldata = c( alldata , c(switchconmean,repeatconmean,switchincmean,repeatincmean))
subject = c( subject , rep(paste("subject",subjectnumber),4))
switchcon = c( switchcon, c( "switch","repeat","switch","repeat"))
congruency= c( congruency, c( "congruent","congruent","incongruent","incongruent"))
subjectnumber = subjectnumber + 1
}
# now put all the data and the conditions in a dataframe named x
x = data.frame( subjectname=subject,
rt=alldata,
switch=switchcon,
con=congruency)
cat("Average switch costs = ",
mean(x[x[,"switch"]=="switch","rt"]) -
mean(x[x[,"switch"]=="repeat","rt"]),"ms\n")
# and do a repeated measure anova
print(summary(aov(rt~switch*con+Error(subjectname/switch*con),data=x)))
}
Mouse and font use
In the following example, you will see how you can use the mouse in your experiments.# File: examples/mouse-and-fonts/mouse.psy # example script doing a choice reaction time experiment # using the mouse, and showing some use of fonts # everything following a hash mark is a comment and is ignored # by the script interpreter options window # experiment runs *not* in full screen escape # you can escape the program bitmapdir bitmaps # location of bitmap files used fontdir fonts # location of font files used centerzero # location 0,0 is center fo screen mouse on # mouse cursor is visible (per default, it is not) render doublebuffer bitmaps # define some bitmap names instruction1 # is in folder bitmaps, instruction1.png instruction2 instruction3 circle phoneoff fonts arialsmall FreeSans.ttf 20 # arialsmall refers to this 20 point size font ariallarge FreeSans.ttf 40 serif FreeSerif.ttf 40 table mousetasktable # table with 4 columns, which can be addressed with @-sign "Click left" -200 100 4 "Click left" -100 200 4 "Click right" 0 250 5 "Click right" 0 300 5 task mousetask table mousetasktable # use this table for choosing conditions font arialsmall # next font to use is arial 20 points show text "ready" 0 0 delay 200 clear 1 # clear first stimulus from screen font ariallarge show text "ready" 0 0 255 0 255 # last three numbers are RedGreenBlue colors delay 200 clear 2 show text @1 # show text, as in column one. delay 500 draw off # hold off showing until all bitmaps positioned show bitmap circle @2 200 # show left bitmap, use x-position from table show bitmap circle @3 200 # show left bitmap, use x-position from table draw on # now show both bitmaps simultaneously readmouse l @4 3000 # wait max 3000 ms until left mouse key is pressed clear 3 clear 4 clear 5 if STATUS != CORRECT show text "error" 0 0 255 0 0 # write in red (colors run from 0 to 255) fi if STATUS == CORRECT font serif # use serif font for "correct" show text "correct" fi delay 1000 clear 6 # clear success or error message save BLOCKNAME STATUS RT MOUSE_X MOUSE_Y # save trial information block mousetaskblock # there is just one block, named "mousetaskblock" pager phoneoff instruction1 instruction2 instruction3 tasklist mousetask 10 # do ten trials end
Wisconsin Card Sorting Test
The Wisconsin Card Sorting Test is based on the work by Grant and Berg, published in 1948. This test is popular in Neuropsychology. The test is designed to measure aspects of mental flexibility; it has similarities with the task-switching paradigm. In this test, the participant must match a card with one of four cards. The difficulty is that it can be matched according to three different things:- The color of the symbols (red/green/blue/yellow).
- The number of the symbols (1 to 4).
- The shape of the symbols (circle,triangle,cross,star).
The participant must figure out which rule to sort to. For example, if the rule is the color rule, cards must be sorted according to the color of the symbols.
To make things more difficult, the rule is changed every ten trials (cards). If the rule gets switched, the participant will obviously make an error. The participant must now figure out what rule to sort to.
And finally, the only feedback the participant get is right or wrong.
The main difference with the task-switching paradigm are: In task-switching paradigms, trial sequences using the same rule are typically shorter, and participants know in advance what rule to apply.
Since there are cheap computers, the WCST has been implemented on computers, and there are, as far as I know, no copyright reasons hindering this (whereas the real card version is distributed and copyrighted).
In this implementation, feedback is given on screen and auditory using sound files.
# File: examples/mouse-and-fonts/wcst/wcst.psy
options
mouse on # mouse is being used, so do not hide it
bitmapdir stimuli # location of the bitmaps
sounddir stimuli # location of sound files
centerzero # center of screen is location 0,0
escape # you can escape by holding escape until end of trial
window
bitmaps
circle1blue # this refers to bitmaps/circle1blue.png
circle1green # etc.
circle1red # each card is 100x100px
circle1yellow # you can change this, of course, making
circle2blue # changes to the SVG file
circle2green
circle2red
circle2yellow
circle3blue
circle3green
circle3red
circle3yellow
circle4blue
circle4green
circle4red
circle4yellow
cross1blue
cross1green
cross1red
cross1yellow
cross2blue
cross2green
cross2red
cross2yellow
cross3blue
cross3green
cross3red
cross3yellow
cross4blue
cross4green
cross4red
cross4yellow
star1blue
star1green
star1red
star1yellow
star2blue
star2green
star2red
star2yellow
star3blue
star3green
star3red
star3yellow
star4blue
star4green
star4red
star4yellow
triangle1blue
triangle1green
triangle1red
triangle1yellow
triangle2blue
triangle2green
triangle2red
triangle2yellow
triangle3blue
triangle3green
triangle3red
triangle3yellow
triangle4blue
triangle4green
triangle4red
triangle4yellow
correct
error
instruction1
instruction2
instruction3
timeout
sounds
good good.wav # this sound file is taken from gnomebaker
wrong wrong.wav # this sound file is taken is from tuxcart
# create table with wcst.r and include here
# one line consists of the following information
# column 1 : card
# column 2 : response (bitmap to be clicked 1 to 4)
# column 3 : trial number in a task sequence, 1 is first, thus rule switch
# column 4 : name of the task
table wcsttable
circle1blue 1 1 "shape" "circle 1 blue"
cross4yellow 3 2 "shape" "cross 4 yellow"
star3yellow 4 3 "shape" "star 3 yellow"
triangle2blue 2 4 "shape" "triangle 2 blue"
circle4green 1 5 "shape" "circle 4 green"
triangle2red 2 6 "shape" "triangle 2 red"
cross2blue 3 7 "shape" "cross 2 blue"
circle4red 1 8 "shape" "circle 4 red"
triangle3red 2 9 "shape" "triangle 3 red"
triangle4yellow 2 10 "shape" "triangle 4 yellow"
circle3green 3 1 "number" "circle 3 green"
circle1green 1 2 "number" "circle 1 green"
triangle3green 3 3 "number" "triangle 3 green"
star4blue 4 4 "number" "star 4 blue"
triangle1red 1 5 "number" "triangle 1 red"
triangle3yellow 3 6 "number" "triangle 3 yellow"
circle4blue 4 7 "number" "circle 4 blue"
star1blue 1 8 "number" "star 1 blue"
star1green 1 9 "number" "star 1 green"
cross2yellow 2 10 "number" "cross 2 yellow"
cross1green 2 1 "color" "cross 1 green"
star1yellow 4 2 "color" "star 1 yellow"
star2yellow 4 3 "color" "star 2 yellow"
triangle1yellow 4 4 "color" "triangle 1 yellow"
circle2yellow 4 5 "color" "circle 2 yellow"
cross3red 1 6 "color" "cross 3 red"
cross4red 1 7 "color" "cross 4 red"
cross4blue 3 8 "color" "cross 4 blue"
star2red 1 9 "color" "star 2 red"
star4red 1 10 "color" "star 4 red"
triangle2yellow 2 1 "shape" "triangle 2 yellow"
star3red 4 2 "shape" "star 3 red"
cross2red 3 3 "shape" "cross 2 red"
triangle4red 2 4 "shape" "triangle 4 red"
circle2green 1 5 "shape" "circle 2 green"
star3blue 4 6 "shape" "star 3 blue"
circle3red 1 7 "shape" "circle 3 red"
cross3green 3 8 "shape" "cross 3 green"
cross1red 3 9 "shape" "cross 1 red"
circle2blue 1 10 "shape" "circle 2 blue"
star1red 1 1 "number" "star 1 red"
cross1yellow 1 2 "number" "cross 1 yellow"
circle4yellow 4 3 "number" "circle 4 yellow"
star3green 3 4 "number" "star 3 green"
triangle3blue 3 5 "number" "triangle 3 blue"
cross3yellow 3 6 "number" "cross 3 yellow"
circle2red 2 7 "number" "circle 2 red"
triangle4blue 4 8 "number" "triangle 4 blue"
cross2green 2 9 "number" "cross 2 green"
circle3blue 3 10 "number" "circle 3 blue"
circle3yellow 4 1 "color" "circle 3 yellow"
star2green 2 2 "color" "star 2 green"
cross1blue 3 3 "color" "cross 1 blue"
triangle1green 2 4 "color" "triangle 1 green"
triangle1blue 3 5 "color" "triangle 1 blue"
triangle4green 2 6 "color" "triangle 4 green"
circle1yellow 4 7 "color" "circle 1 yellow"
star2blue 3 8 "color" "star 2 blue"
star4green 2 9 "color" "star 4 green"
cross4green 2 10 "color" "cross 4 green"
task wcst
table wcsttable # use the table wcsttable
delay 1000 # wait 1 second
draw off # show next 5 bitmaps at once
show bitmap circle1red -175 -100 # bitmap number 1
show bitmap triangle2green -25 -100 # bitmap number 2
show bitmap cross3blue 125 -100 # bitmap number 3
show bitmap star4yellow 275 -100 # bitmap number 4
show bitmap @1 -300 200 # bitmap number 5
draw on # now show them
set $a 0 # once clicked, $a will be clicked-bitmap number
readmouse l @2 10000 range 1 4 # wait for left mouse click on rect 1-4 for 10sec
set $a bitmap-under-mouse MOUSE_X MOUSE_Y # which bitmap was clicked?
clear 5 # erase the last bitmap from screen
if $a == 1 # if bitmap 1 was clicked, set variable newx to -175
set $newx -175
fi # end of if statement
if $a == 2
set $newx -25
fi
if $a == 3
set $newx 125
fi
if $a == 4
set $newx 275
fi
if $a == 0 # this means, nothing clicked, therefor timeout
show bitmap timeout -300 25 # show the timeout message
fi
if $a > 0
show bitmap @1 $newx 25 # show the same card (6) underneath the one clicked
fi
delay 500 # keep it for 500 ms
if STATUS == CORRECT # if match was correct
sound good # give vocal feedback
show bitmap correct $newx 100 # show message "correct", bitmap 7
delay 200
clear -1
delay 200
show bitmap correct $newx 100 # show message "correct", bitmap 8
fi
if STATUS != CORRECT # if match was incorrect
sound wrong # give vocal feedback
show bitmap error $newx 100 # show message "error", bitmap 7
delay 200
clear -1
delay 200
show bitmap error $newx 100 # show message "error", bitmap 8
fi
delay 1000 # wait a second for feedback to be read/heard
clear 6 7 # clear feedback card (6) and feedback message (7)
save @1 @2 @3 @4 @5 RT STATUS $a # save trial information to disk
block wcstblock # there is just one block. Name it "wcstblock"
pager instruction1 instruction2 instruction3
tasklist
wcst 60 fixed # 60 trials, fixed follows order of table, is essential
end
system R CMD BATCH feedback.r
bitmap_from_file feedback.png
wait_for_key
Attentional blink paradigm
The attentional blink paradigm is based on a study published in the journal Nature in 1994 by Duncan, Ward, and Shapiro. The study, titled "Direct measurement of attentional dwell time in human vision" shows a difficulty to decide whether an object matches a predefined target when it follows another attended object within 100-600 ms.The task explained here is based on experiment 2 of the aforementioned study. There are a few differences, but the idea is the same.
There are many different conditions, resulting from the various locations stimuli can appear and the intervals between the two stimuli. In the example below, there are many conditions. In the example, the conditions are all described in a table. The table can in principle be produced by hand, but that is a lot of work. It can easily be produced with an R script (available in downloadable package), which just loops through the different conditions and writes a line for each condition (how to do this is in the examples section of the downloadable psytoolkit distribution).
Attentional blink paradigm with table
# File: examples/attentional-blink/ab.psy
# attentional blink
# based on experiment 2 of Duncan, Ward & Shapiro (1994) in Nature.
options
window
bitmapdir stimuli # location of the bitmaps
centerzero # center of screen is location 0,0
escape # you can escape by holding escape until end of trial
render doublebuffer
bitmaps
t1 # target
n1 # nontarget1
n2 # nontarget2
n3 # nontarget3
correct # correct feedback message
error # error feedback message
instruction1 # instruction screen 1
instruction2 # instruction screen 2
box # box stimulus appear in
mask # mask
fix # fixation point
help # gives help following error
table abtable # table created with the R file ab-table.r
0 n1 t1 -200 -200 1 "0 n1 t1 -200 -200 1"
100 n1 t1 -200 -200 1 "100 n1 t1 -200 -200 1"
100 t1 n1 -200 -200 1 "-100 t1 n1 -200 -200 1"
196 n1 t1 -200 -200 1 "196 n1 t1 -200 -200 1"
196 t1 n1 -200 -200 1 "-196 t1 n1 -200 -200 1"
280 n1 t1 -200 -200 1 "280 n1 t1 -200 -200 1"
280 t1 n1 -200 -200 1 "-280 t1 n1 -200 -200 1"
430 n1 t1 -200 -200 1 "430 n1 t1 -200 -200 1"
430 t1 n1 -200 -200 1 "-430 t1 n1 -200 -200 1"
590 n1 t1 -200 -200 1 "590 n1 t1 -200 -200 1"
590 t1 n1 -200 -200 1 "-590 t1 n1 -200 -200 1"
900 n1 t1 -200 -200 1 "900 n1 t1 -200 -200 1"
900 t1 n1 -200 -200 1 "-900 t1 n1 -200 -200 1"
0 n1 t1 -200 200 1 "0 n1 t1 -200 200 1"
100 n1 t1 -200 200 1 "100 n1 t1 -200 200 1"
100 t1 n1 -200 200 1 "-100 t1 n1 -200 200 1"
196 n1 t1 -200 200 1 "196 n1 t1 -200 200 1"
196 t1 n1 -200 200 1 "-196 t1 n1 -200 200 1"
280 n1 t1 -200 200 1 "280 n1 t1 -200 200 1"
280 t1 n1 -200 200 1 "-280 t1 n1 -200 200 1"
430 n1 t1 -200 200 1 "430 n1 t1 -200 200 1"
430 t1 n1 -200 200 1 "-430 t1 n1 -200 200 1"
590 n1 t1 -200 200 1 "590 n1 t1 -200 200 1"
590 t1 n1 -200 200 1 "-590 t1 n1 -200 200 1"
900 n1 t1 -200 200 1 "900 n1 t1 -200 200 1"
900 t1 n1 -200 200 1 "-900 t1 n1 -200 200 1"
0 n1 t1 200 -200 1 "0 n1 t1 200 -200 1"
100 n1 t1 200 -200 1 "100 n1 t1 200 -200 1"
100 t1 n1 200 -200 1 "-100 t1 n1 200 -200 1"
196 n1 t1 200 -200 1 "196 n1 t1 200 -200 1"
196 t1 n1 200 -200 1 "-196 t1 n1 200 -200 1"
280 n1 t1 200 -200 1 "280 n1 t1 200 -200 1"
280 t1 n1 200 -200 1 "-280 t1 n1 200 -200 1"
430 n1 t1 200 -200 1 "430 n1 t1 200 -200 1"
430 t1 n1 200 -200 1 "-430 t1 n1 200 -200 1"
590 n1 t1 200 -200 1 "590 n1 t1 200 -200 1"
590 t1 n1 200 -200 1 "-590 t1 n1 200 -200 1"
900 n1 t1 200 -200 1 "900 n1 t1 200 -200 1"
900 t1 n1 200 -200 1 "-900 t1 n1 200 -200 1"
0 n1 t1 200 200 1 "0 n1 t1 200 200 1"
100 n1 t1 200 200 1 "100 n1 t1 200 200 1"
100 t1 n1 200 200 1 "-100 t1 n1 200 200 1"
196 n1 t1 200 200 1 "196 n1 t1 200 200 1"
196 t1 n1 200 200 1 "-196 t1 n1 200 200 1"
280 n1 t1 200 200 1 "280 n1 t1 200 200 1"
280 t1 n1 200 200 1 "-280 t1 n1 200 200 1"
430 n1 t1 200 200 1 "430 n1 t1 200 200 1"
430 t1 n1 200 200 1 "-430 t1 n1 200 200 1"
590 n1 t1 200 200 1 "590 n1 t1 200 200 1"
590 t1 n1 200 200 1 "-590 t1 n1 200 200 1"
900 n1 t1 200 200 1 "900 n1 t1 200 200 1"
900 t1 n1 200 200 1 "-900 t1 n1 200 200 1"
0 n2 t1 -200 -200 1 "0 n2 t1 -200 -200 1"
100 n2 t1 -200 -200 1 "100 n2 t1 -200 -200 1"
100 t1 n2 -200 -200 1 "-100 t1 n2 -200 -200 1"
196 n2 t1 -200 -200 1 "196 n2 t1 -200 -200 1"
196 t1 n2 -200 -200 1 "-196 t1 n2 -200 -200 1"
280 n2 t1 -200 -200 1 "280 n2 t1 -200 -200 1"
280 t1 n2 -200 -200 1 "-280 t1 n2 -200 -200 1"
430 n2 t1 -200 -200 1 "430 n2 t1 -200 -200 1"
430 t1 n2 -200 -200 1 "-430 t1 n2 -200 -200 1"
590 n2 t1 -200 -200 1 "590 n2 t1 -200 -200 1"
590 t1 n2 -200 -200 1 "-590 t1 n2 -200 -200 1"
900 n2 t1 -200 -200 1 "900 n2 t1 -200 -200 1"
900 t1 n2 -200 -200 1 "-900 t1 n2 -200 -200 1"
0 n2 t1 -200 200 1 "0 n2 t1 -200 200 1"
100 n2 t1 -200 200 1 "100 n2 t1 -200 200 1"
100 t1 n2 -200 200 1 "-100 t1 n2 -200 200 1"
196 n2 t1 -200 200 1 "196 n2 t1 -200 200 1"
196 t1 n2 -200 200 1 "-196 t1 n2 -200 200 1"
280 n2 t1 -200 200 1 "280 n2 t1 -200 200 1"
280 t1 n2 -200 200 1 "-280 t1 n2 -200 200 1"
430 n2 t1 -200 200 1 "430 n2 t1 -200 200 1"
430 t1 n2 -200 200 1 "-430 t1 n2 -200 200 1"
590 n2 t1 -200 200 1 "590 n2 t1 -200 200 1"
590 t1 n2 -200 200 1 "-590 t1 n2 -200 200 1"
900 n2 t1 -200 200 1 "900 n2 t1 -200 200 1"
900 t1 n2 -200 200 1 "-900 t1 n2 -200 200 1"
0 n2 t1 200 -200 1 "0 n2 t1 200 -200 1"
100 n2 t1 200 -200 1 "100 n2 t1 200 -200 1"
100 t1 n2 200 -200 1 "-100 t1 n2 200 -200 1"
196 n2 t1 200 -200 1 "196 n2 t1 200 -200 1"
196 t1 n2 200 -200 1 "-196 t1 n2 200 -200 1"
280 n2 t1 200 -200 1 "280 n2 t1 200 -200 1"
280 t1 n2 200 -200 1 "-280 t1 n2 200 -200 1"
430 n2 t1 200 -200 1 "430 n2 t1 200 -200 1"
430 t1 n2 200 -200 1 "-430 t1 n2 200 -200 1"
590 n2 t1 200 -200 1 "590 n2 t1 200 -200 1"
590 t1 n2 200 -200 1 "-590 t1 n2 200 -200 1"
900 n2 t1 200 -200 1 "900 n2 t1 200 -200 1"
900 t1 n2 200 -200 1 "-900 t1 n2 200 -200 1"
0 n2 t1 200 200 1 "0 n2 t1 200 200 1"
100 n2 t1 200 200 1 "100 n2 t1 200 200 1"
100 t1 n2 200 200 1 "-100 t1 n2 200 200 1"
196 n2 t1 200 200 1 "196 n2 t1 200 200 1"
196 t1 n2 200 200 1 "-196 t1 n2 200 200 1"
280 n2 t1 200 200 1 "280 n2 t1 200 200 1"
280 t1 n2 200 200 1 "-280 t1 n2 200 200 1"
430 n2 t1 200 200 1 "430 n2 t1 200 200 1"
430 t1 n2 200 200 1 "-430 t1 n2 200 200 1"
590 n2 t1 200 200 1 "590 n2 t1 200 200 1"
590 t1 n2 200 200 1 "-590 t1 n2 200 200 1"
900 n2 t1 200 200 1 "900 n2 t1 200 200 1"
900 t1 n2 200 200 1 "-900 t1 n2 200 200 1"
0 n3 t1 -200 -200 1 "0 n3 t1 -200 -200 1"
100 n3 t1 -200 -200 1 "100 n3 t1 -200 -200 1"
100 t1 n3 -200 -200 1 "-100 t1 n3 -200 -200 1"
196 n3 t1 -200 -200 1 "196 n3 t1 -200 -200 1"
196 t1 n3 -200 -200 1 "-196 t1 n3 -200 -200 1"
280 n3 t1 -200 -200 1 "280 n3 t1 -200 -200 1"
280 t1 n3 -200 -200 1 "-280 t1 n3 -200 -200 1"
430 n3 t1 -200 -200 1 "430 n3 t1 -200 -200 1"
430 t1 n3 -200 -200 1 "-430 t1 n3 -200 -200 1"
590 n3 t1 -200 -200 1 "590 n3 t1 -200 -200 1"
590 t1 n3 -200 -200 1 "-590 t1 n3 -200 -200 1"
900 n3 t1 -200 -200 1 "900 n3 t1 -200 -200 1"
900 t1 n3 -200 -200 1 "-900 t1 n3 -200 -200 1"
0 n3 t1 -200 200 1 "0 n3 t1 -200 200 1"
100 n3 t1 -200 200 1 "100 n3 t1 -200 200 1"
100 t1 n3 -200 200 1 "-100 t1 n3 -200 200 1"
196 n3 t1 -200 200 1 "196 n3 t1 -200 200 1"
196 t1 n3 -200 200 1 "-196 t1 n3 -200 200 1"
280 n3 t1 -200 200 1 "280 n3 t1 -200 200 1"
280 t1 n3 -200 200 1 "-280 t1 n3 -200 200 1"
430 n3 t1 -200 200 1 "430 n3 t1 -200 200 1"
430 t1 n3 -200 200 1 "-430 t1 n3 -200 200 1"
590 n3 t1 -200 200 1 "590 n3 t1 -200 200 1"
590 t1 n3 -200 200 1 "-590 t1 n3 -200 200 1"
900 n3 t1 -200 200 1 "900 n3 t1 -200 200 1"
900 t1 n3 -200 200 1 "-900 t1 n3 -200 200 1"
0 n3 t1 200 -200 1 "0 n3 t1 200 -200 1"
100 n3 t1 200 -200 1 "100 n3 t1 200 -200 1"
100 t1 n3 200 -200 1 "-100 t1 n3 200 -200 1"
196 n3 t1 200 -200 1 "196 n3 t1 200 -200 1"
196 t1 n3 200 -200 1 "-196 t1 n3 200 -200 1"
280 n3 t1 200 -200 1 "280 n3 t1 200 -200 1"
280 t1 n3 200 -200 1 "-280 t1 n3 200 -200 1"
430 n3 t1 200 -200 1 "430 n3 t1 200 -200 1"
430 t1 n3 200 -200 1 "-430 t1 n3 200 -200 1"
590 n3 t1 200 -200 1 "590 n3 t1 200 -200 1"
590 t1 n3 200 -200 1 "-590 t1 n3 200 -200 1"
900 n3 t1 200 -200 1 "900 n3 t1 200 -200 1"
900 t1 n3 200 -200 1 "-900 t1 n3 200 -200 1"
0 n3 t1 200 200 1 "0 n3 t1 200 200 1"
100 n3 t1 200 200 1 "100 n3 t1 200 200 1"
100 t1 n3 200 200 1 "-100 t1 n3 200 200 1"
196 n3 t1 200 200 1 "196 n3 t1 200 200 1"
196 t1 n3 200 200 1 "-196 t1 n3 200 200 1"
280 n3 t1 200 200 1 "280 n3 t1 200 200 1"
280 t1 n3 200 200 1 "-280 t1 n3 200 200 1"
430 n3 t1 200 200 1 "430 n3 t1 200 200 1"
430 t1 n3 200 200 1 "-430 t1 n3 200 200 1"
590 n3 t1 200 200 1 "590 n3 t1 200 200 1"
590 t1 n3 200 200 1 "-590 t1 n3 200 200 1"
900 n3 t1 200 200 1 "900 n3 t1 200 200 1"
900 t1 n3 200 200 1 "-900 t1 n3 200 200 1"
0 n1 n2 -200 -200 2 "0 n1 n2 -200 -200 2"
100 n1 n2 -200 -200 2 "100 n1 n2 -200 -200 2"
100 n2 n1 -200 -200 2 "-100 n2 n1 -200 -200 2"
196 n1 n2 -200 -200 2 "196 n1 n2 -200 -200 2"
196 n2 n1 -200 -200 2 "-196 n2 n1 -200 -200 2"
280 n1 n2 -200 -200 2 "280 n1 n2 -200 -200 2"
280 n2 n1 -200 -200 2 "-280 n2 n1 -200 -200 2"
430 n1 n2 -200 -200 2 "430 n1 n2 -200 -200 2"
430 n2 n1 -200 -200 2 "-430 n2 n1 -200 -200 2"
590 n1 n2 -200 -200 2 "590 n1 n2 -200 -200 2"
590 n2 n1 -200 -200 2 "-590 n2 n1 -200 -200 2"
900 n1 n2 -200 -200 2 "900 n1 n2 -200 -200 2"
900 n2 n1 -200 -200 2 "-900 n2 n1 -200 -200 2"
0 n1 n2 -200 200 2 "0 n1 n2 -200 200 2"
100 n1 n2 -200 200 2 "100 n1 n2 -200 200 2"
100 n2 n1 -200 200 2 "-100 n2 n1 -200 200 2"
196 n1 n2 -200 200 2 "196 n1 n2 -200 200 2"
196 n2 n1 -200 200 2 "-196 n2 n1 -200 200 2"
280 n1 n2 -200 200 2 "280 n1 n2 -200 200 2"
280 n2 n1 -200 200 2 "-280 n2 n1 -200 200 2"
430 n1 n2 -200 200 2 "430 n1 n2 -200 200 2"
430 n2 n1 -200 200 2 "-430 n2 n1 -200 200 2"
590 n1 n2 -200 200 2 "590 n1 n2 -200 200 2"
590 n2 n1 -200 200 2 "-590 n2 n1 -200 200 2"
900 n1 n2 -200 200 2 "900 n1 n2 -200 200 2"
900 n2 n1 -200 200 2 "-900 n2 n1 -200 200 2"
0 n1 n2 200 -200 2 "0 n1 n2 200 -200 2"
100 n1 n2 200 -200 2 "100 n1 n2 200 -200 2"
100 n2 n1 200 -200 2 "-100 n2 n1 200 -200 2"
196 n1 n2 200 -200 2 "196 n1 n2 200 -200 2"
196 n2 n1 200 -200 2 "-196 n2 n1 200 -200 2"
280 n1 n2 200 -200 2 "280 n1 n2 200 -200 2"
280 n2 n1 200 -200 2 "-280 n2 n1 200 -200 2"
430 n1 n2 200 -200 2 "430 n1 n2 200 -200 2"
430 n2 n1 200 -200 2 "-430 n2 n1 200 -200 2"
590 n1 n2 200 -200 2 "590 n1 n2 200 -200 2"
590 n2 n1 200 -200 2 "-590 n2 n1 200 -200 2"
900 n1 n2 200 -200 2 "900 n1 n2 200 -200 2"
900 n2 n1 200 -200 2 "-900 n2 n1 200 -200 2"
0 n1 n2 200 200 2 "0 n1 n2 200 200 2"
100 n1 n2 200 200 2 "100 n1 n2 200 200 2"
100 n2 n1 200 200 2 "-100 n2 n1 200 200 2"
196 n1 n2 200 200 2 "196 n1 n2 200 200 2"
196 n2 n1 200 200 2 "-196 n2 n1 200 200 2"
280 n1 n2 200 200 2 "280 n1 n2 200 200 2"
280 n2 n1 200 200 2 "-280 n2 n1 200 200 2"
430 n1 n2 200 200 2 "430 n1 n2 200 200 2"
430 n2 n1 200 200 2 "-430 n2 n1 200 200 2"
590 n1 n2 200 200 2 "590 n1 n2 200 200 2"
590 n2 n1 200 200 2 "-590 n2 n1 200 200 2"
900 n1 n2 200 200 2 "900 n1 n2 200 200 2"
900 n2 n1 200 200 2 "-900 n2 n1 200 200 2"
0 n1 n3 -200 -200 2 "0 n1 n3 -200 -200 2"
100 n1 n3 -200 -200 2 "100 n1 n3 -200 -200 2"
100 n3 n1 -200 -200 2 "-100 n3 n1 -200 -200 2"
196 n1 n3 -200 -200 2 "196 n1 n3 -200 -200 2"
196 n3 n1 -200 -200 2 "-196 n3 n1 -200 -200 2"
280 n1 n3 -200 -200 2 "280 n1 n3 -200 -200 2"
280 n3 n1 -200 -200 2 "-280 n3 n1 -200 -200 2"
430 n1 n3 -200 -200 2 "430 n1 n3 -200 -200 2"
430 n3 n1 -200 -200 2 "-430 n3 n1 -200 -200 2"
590 n1 n3 -200 -200 2 "590 n1 n3 -200 -200 2"
590 n3 n1 -200 -200 2 "-590 n3 n1 -200 -200 2"
900 n1 n3 -200 -200 2 "900 n1 n3 -200 -200 2"
900 n3 n1 -200 -200 2 "-900 n3 n1 -200 -200 2"
0 n1 n3 -200 200 2 "0 n1 n3 -200 200 2"
100 n1 n3 -200 200 2 "100 n1 n3 -200 200 2"
100 n3 n1 -200 200 2 "-100 n3 n1 -200 200 2"
196 n1 n3 -200 200 2 "196 n1 n3 -200 200 2"
196 n3 n1 -200 200 2 "-196 n3 n1 -200 200 2"
280 n1 n3 -200 200 2 "280 n1 n3 -200 200 2"
280 n3 n1 -200 200 2 "-280 n3 n1 -200 200 2"
430 n1 n3 -200 200 2 "430 n1 n3 -200 200 2"
430 n3 n1 -200 200 2 "-430 n3 n1 -200 200 2"
590 n1 n3 -200 200 2 "590 n1 n3 -200 200 2"
590 n3 n1 -200 200 2 "-590 n3 n1 -200 200 2"
900 n1 n3 -200 200 2 "900 n1 n3 -200 200 2"
900 n3 n1 -200 200 2 "-900 n3 n1 -200 200 2"
0 n1 n3 200 -200 2 "0 n1 n3 200 -200 2"
100 n1 n3 200 -200 2 "100 n1 n3 200 -200 2"
100 n3 n1 200 -200 2 "-100 n3 n1 200 -200 2"
196 n1 n3 200 -200 2 "196 n1 n3 200 -200 2"
196 n3 n1 200 -200 2 "-196 n3 n1 200 -200 2"
280 n1 n3 200 -200 2 "280 n1 n3 200 -200 2"
280 n3 n1 200 -200 2 "-280 n3 n1 200 -200 2"
430 n1 n3 200 -200 2 "430 n1 n3 200 -200 2"
430 n3 n1 200 -200 2 "-430 n3 n1 200 -200 2"
590 n1 n3 200 -200 2 "590 n1 n3 200 -200 2"
590 n3 n1 200 -200 2 "-590 n3 n1 200 -200 2"
900 n1 n3 200 -200 2 "900 n1 n3 200 -200 2"
900 n3 n1 200 -200 2 "-900 n3 n1 200 -200 2"
0 n1 n3 200 200 2 "0 n1 n3 200 200 2"
100 n1 n3 200 200 2 "100 n1 n3 200 200 2"
100 n3 n1 200 200 2 "-100 n3 n1 200 200 2"
196 n1 n3 200 200 2 "196 n1 n3 200 200 2"
196 n3 n1 200 200 2 "-196 n3 n1 200 200 2"
280 n1 n3 200 200 2 "280 n1 n3 200 200 2"
280 n3 n1 200 200 2 "-280 n3 n1 200 200 2"
430 n1 n3 200 200 2 "430 n1 n3 200 200 2"
430 n3 n1 200 200 2 "-430 n3 n1 200 200 2"
590 n1 n3 200 200 2 "590 n1 n3 200 200 2"
590 n3 n1 200 200 2 "-590 n3 n1 200 200 2"
900 n1 n3 200 200 2 "900 n1 n3 200 200 2"
900 n3 n1 200 200 2 "-900 n3 n1 200 200 2"
0 n2 n3 -200 -200 2 "0 n2 n3 -200 -200 2"
100 n2 n3 -200 -200 2 "100 n2 n3 -200 -200 2"
100 n3 n2 -200 -200 2 "-100 n3 n2 -200 -200 2"
196 n2 n3 -200 -200 2 "196 n2 n3 -200 -200 2"
196 n3 n2 -200 -200 2 "-196 n3 n2 -200 -200 2"
280 n2 n3 -200 -200 2 "280 n2 n3 -200 -200 2"
280 n3 n2 -200 -200 2 "-280 n3 n2 -200 -200 2"
430 n2 n3 -200 -200 2 "430 n2 n3 -200 -200 2"
430 n3 n2 -200 -200 2 "-430 n3 n2 -200 -200 2"
590 n2 n3 -200 -200 2 "590 n2 n3 -200 -200 2"
590 n3 n2 -200 -200 2 "-590 n3 n2 -200 -200 2"
900 n2 n3 -200 -200 2 "900 n2 n3 -200 -200 2"
900 n3 n2 -200 -200 2 "-900 n3 n2 -200 -200 2"
0 n2 n3 -200 200 2 "0 n2 n3 -200 200 2"
100 n2 n3 -200 200 2 "100 n2 n3 -200 200 2"
100 n3 n2 -200 200 2 "-100 n3 n2 -200 200 2"
196 n2 n3 -200 200 2 "196 n2 n3 -200 200 2"
196 n3 n2 -200 200 2 "-196 n3 n2 -200 200 2"
280 n2 n3 -200 200 2 "280 n2 n3 -200 200 2"
280 n3 n2 -200 200 2 "-280 n3 n2 -200 200 2"
430 n2 n3 -200 200 2 "430 n2 n3 -200 200 2"
430 n3 n2 -200 200 2 "-430 n3 n2 -200 200 2"
590 n2 n3 -200 200 2 "590 n2 n3 -200 200 2"
590 n3 n2 -200 200 2 "-590 n3 n2 -200 200 2"
900 n2 n3 -200 200 2 "900 n2 n3 -200 200 2"
900 n3 n2 -200 200 2 "-900 n3 n2 -200 200 2"
0 n2 n3 200 -200 2 "0 n2 n3 200 -200 2"
100 n2 n3 200 -200 2 "100 n2 n3 200 -200 2"
100 n3 n2 200 -200 2 "-100 n3 n2 200 -200 2"
196 n2 n3 200 -200 2 "196 n2 n3 200 -200 2"
196 n3 n2 200 -200 2 "-196 n3 n2 200 -200 2"
280 n2 n3 200 -200 2 "280 n2 n3 200 -200 2"
280 n3 n2 200 -200 2 "-280 n3 n2 200 -200 2"
430 n2 n3 200 -200 2 "430 n2 n3 200 -200 2"
430 n3 n2 200 -200 2 "-430 n3 n2 200 -200 2"
590 n2 n3 200 -200 2 "590 n2 n3 200 -200 2"
590 n3 n2 200 -200 2 "-590 n3 n2 200 -200 2"
900 n2 n3 200 -200 2 "900 n2 n3 200 -200 2"
900 n3 n2 200 -200 2 "-900 n3 n2 200 -200 2"
0 n2 n3 200 200 2 "0 n2 n3 200 200 2"
100 n2 n3 200 200 2 "100 n2 n3 200 200 2"
100 n3 n2 200 200 2 "-100 n3 n2 200 200 2"
196 n2 n3 200 200 2 "196 n2 n3 200 200 2"
196 n3 n2 200 200 2 "-196 n3 n2 200 200 2"
280 n2 n3 200 200 2 "280 n2 n3 200 200 2"
280 n3 n2 200 200 2 "-280 n3 n2 200 200 2"
430 n2 n3 200 200 2 "430 n2 n3 200 200 2"
430 n3 n2 200 200 2 "-430 n3 n2 200 200 2"
590 n2 n3 200 200 2 "590 n2 n3 200 200 2"
590 n3 n2 200 200 2 "-590 n3 n2 200 200 2"
900 n2 n3 200 200 2 "900 n2 n3 200 200 2"
900 n3 n2 200 200 2 "-900 n3 n2 200 200 2"
task ab
table abtable # use the table abtable
keys lshift rshift space # keys to be used
draw off # hold of before showing next 5 bitmaps
show bitmap fix # draw fix point and 4 boxes
show bitmap box -200 0
show bitmap box 200 0
show bitmap box 0 200
show bitmap box 0 -200
draw on # show them now all at the same time
delay 500 # and wait for 500 ms
readkey 1 120000 # wait up to 2 minutes for any key to start task
delay 1000
show bitmap @2 @4 0 # show stimulus 1 (left or right)
delay 50 # presentation time of stimulus 1
show bitmap mask @4 0 # show mask over stimulus 1
delay @1 # delay between stimuli (which is shorter than SOA)
show bitmap @3 0 @5 # show stimlus 2 (top or bottom)
delay 50 # presentation time of stimulus 2
show bitmap mask 0 @5 # show mask over stimulus 1
readkey @6 5000 # wait for response up to 5s
clear 1 # erase fixpoint
if STATUS == CORRECT # what if correct key was pressed
show bitmap correct # show the correct feedback stimulus
delay 500 # for 500 ms
clear -1 # and erase that stimulus from the screen
fi # fi is end of if block
if STATUS != CORRECT # what if wrong key was pressed
show bitmap error # show the error feedback stimulus
delay 500 # for 500 ms
clear -1 # and erase that stimulus from the screen
fi # fi is end of if block
delay 500 # wait 500 ms
clear 2 3 4 5 6 7 8 9 # clear all remaining stimuli
save @7 RT STATUS # save trial information to disk
block instructionblock
pager instruction1 instruction2 # show instructions/browse
block abblock 20
tasklist
ab 10
end
system R CMD BATCH feedback.r
bitmap_from_file feedback.png
wait_for_key
As you can easily see, the task description is much shorter than the table. The table is the big part of an experimental paradigm. And when there are many experimental variables, you can get a long table. In principle, you could use a different coding strategy, in which you choose certain experimental variables using variables. You could even completely get rid of the table.
Multiple Object Tracking Task
Multiple Object Tracking is a difficult task, especially if all the objects that are moving around are looking the same. Read more here: (click).Multiple object tracking is relatively easy to implement with PsyToolkit. Below you see what the example experiment (code below) looks like. With PsyToolkit you can create game-like experiments that are especially attractive for work with children.
Have a look at the examples folder, which contains two examples, including a very simple example of object tracking (way too simple for a real experiment) below here:
Simple object tracking experiment
# File: examples/object-tracking/object-tracking-simple.psy
options
window
escape
mouse on
centerzero
transparency on
render doublebuffer
bitmaps
backgroundpicture
red
green
error
success
simpleinstructions
start
task trackobjects
show bitmap backgroundpicture
mouse show
show bitmap start
readmouse l 1 100000
clear 2
mouse hide # during movement, do not show the mouse
set $x1 random -300 -50 # choose random x/y positions
set $x2 random -300 -50 # of four sprites
set $x3 random 50 300
set $x4 random 50 300
set $y1 random -250 -50
set $y2 random 50 250
set $y3 random -250 -50
set $y4 random 50 250
sprite create green $x1 $y1 # and now apply these here
sprite create red $x2 $y2
sprite create red $x3 $y4
sprite create red $x4 $y4
sprites bounce all # sprites will bounce
sprite 1 move direction 30 3 # 30 is angle and 3 is speed
sprite 2 move direction 60 3
sprite 3 move direction 10 3
sprite 4 move direction 25 3
sprites display all # now that they are set up, show them
delay 1000 # move around for 1 second
sprite 1 bitmap red # turn sprite #1 red like the others
delay 5000 # keep moving for 5 secs
sprites freeze all # stop moving
mouse show # show the mouse again to enable clicking
readmouse_sprite l 1 5000 # wait for clicking a sprite
if STATUS == CORRECT
show bitmap success
fi
if STATUS != CORRECT
show bitmap error
fi
sprite 1 bitmap green # show where sprite one was
delay 1500
sprite 1 hide # hide the sprites
sprite 2 hide
sprite 3 hide
sprite 4 hide
clear 3
sprites delete all # you must delete them, otherwise they will stay
clear 1
save RT STATUS
######################################################################
message simpleinstructions
block firstblock
tasklist
trackobjects 10
end